THE DNA
DNA is a long polymer of deoxyribonucleotides. The length of DNA is
usually defined as number of nucleotides (or a pair of nucleotide referred
to as base pairs) present in it. This also is the characteristic of an organism.
For example, a bacteriophage known as f ×174 has 5386 nucleotides,
Bacteriophage lambda has 48502 base pairs (bp), Escherichia coli has
4.6 × 106 bp, and haploid content of human DNA is 3.3 × 109 bp. Let us
discuss the structure of such a long polymer.
Structure of Polynucleotide Chain
Let us recapitulate the chemical structure of a polynucleotide chain (DNA
or RNA). A nucleotide has three components – a nitrogenous base, a
pentose sugar (ribose in case of RNA, and deoxyribose for DNA), and a
phosphate group. There are two types of nitrogenous bases – Purines
(Adenine and Guanine), and Pyrimidines (Cytosine, Uracil and Thymine).
Cytosine is common for both DNA and RNA and Thymine is present in
DNA. Uracil is present in RNA at the place of Thymine. A nitrogenous
base is linked to the OH of 1' C pentose sugar through a N-glycosidic
linkage to form a nucleoside, such as adenosine or deoxyadenosine,
guanosine or deoxyguanosine, cytidine or deoxycytidine and uridine or
deoxythymidine. When a phosphate group is linked to OH of 5' C of a
nucleoside through phosphoester linkage, a corresponding nucleotide
(or deoxynucleotide depending upon the type of sugar present) is formed.
Two nucleotides are linked through 3'-5' phosphodiester linkage to form
a dinucleotide. More nucleotides can be joined in such a manner to form
a polynucleotide chain. A polymer thus formed has at one end a freephosphate moiety at 5' -end of sugar, which is referred to as 5’-end of
polynucleotide chain. Similarly, at the other end of the polymer the sugar
has a free OH of 3'C group which is referred to as 3' -end of the
polynucleotide chain. The backbone of a polynucleotide chain is formed
due to sugar and phosphates. The nitrogenous bases linked to sugar
moiety project from the backbone (Figure-1)
In RNA, every nucleotide residue has an additional –OH group present
at 2' -position in the ribose. Also, in RNA the uracil is found at the place of
thymine (5-methyl uracil, another chemical name for thymine).
DNA as an acidic substance present in nucleus was first identified by
Friedrich Meischer in 1869. He named it as ‘Nuclein’. However, due to
technical limitation in isolating such a long polymer intact, the elucidation
of structure of DNA remained elusive for a very long period of time. It was
only in 1953 that James Watson and Francis Crick, based on the X-ray
diffraction data produced by Maurice Wilkins and Rosalind Franklin,
proposed a very simple but famous Double Helix model for the structure
of DNA. One of the hallmarks of their proposition was base pairing between
the two strands of polynucleotide chains. However, this proposition was
also based on the observation of Erwin Chargaff that for a double stranded
DNA, the ratios between Adenine and Thymine and Guanine and Cytosine
are constant and equals one.
The base pairing confers a very unique property to the polynucleotide
chains. They are said to be complementary to each other, and therefore if
the sequence of bases in one strand is known then the sequence in other
strand can be predicted. Also, if each strand from a DNA (let us call it as a
parental DNA) acts as a template for synthesis of a new strand, the two
double stranded DNA (let us call them as daughter DNA) thus, produced
would be identical to the parental DNA molecule. Because of this, the genetic
implications of the structure of DNA became very clear.
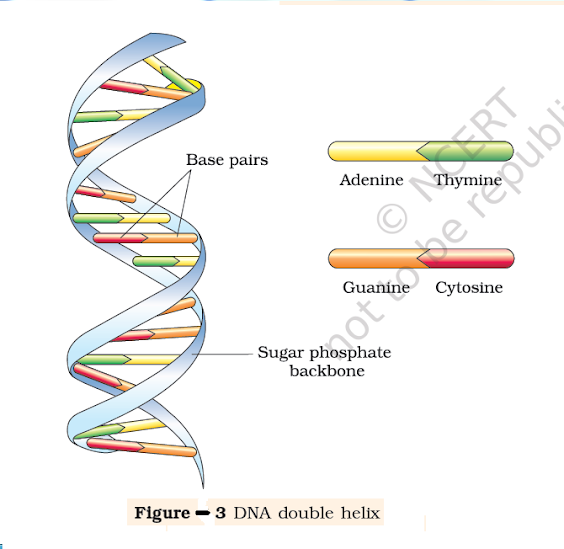
The salient features of the Double-helix structure of DNA are as follows:
(i) It is made of two polynucleotide chains, where the backbone is
constituted by sugar-phosphate, and the bases project inside.
(ii) The two chains have anti-parallel polarity. It means, if one
chain has the polarity 5'>3', the other has 3'>5' .
(iii) The bases in two strands are paired through hydrogen bond
(H-bonds) forming base pairs (bp). Adenine forms two hydrogen
bonds with Thymine from opposite strand and vice-versa.
Similarly, Guanine is bonded with Cytosine with three H-bonds.
As a result, always a purine comes opposite to a pyrimidine. This
generates approximately uniform distance between the two
strands of the helix (Figure-2). (iv) The two chains are coiled in a right-handed fashion. The pitch
of the helix is 3.4 nm (a nanometre is one billionth of a
metre, that is 10-9 m) and there are roughly 10 bp in each
2020-turn. Consequently, the distance
between a bp in a helix is
approximately 0.34 nm.
(iv) The two chains are coiled in a right-handed fashion. The pitch
of the helix is 3.4 nm (a nanometre is one billionth of a
metre, that is 10-9 m) and there are roughly 10 bp in each
2020-turn. Consequently, the distance
between a bp in a helix is
approximately 0.34 nm.
 (iv) The two chains are coiled in a right-handed fashion. The pitch
of the helix is 3.4 nm (a nanometre is one billionth of a
metre, that is 10-9 m) and there are roughly 10 bp in each
2020-turn. Consequently, the distance
between a bp in a helix is
approximately 0.34 nm.
(iv) The two chains are coiled in a right-handed fashion. The pitch
of the helix is 3.4 nm (a nanometre is one billionth of a
metre, that is 10-9 m) and there are roughly 10 bp in each
2020-turn. Consequently, the distance
between a bp in a helix is
approximately 0.34 nm. (v) The plane of one base pair stacks
over the other in double helix. This,
in addition to H-bonds, confers
stability of the helical structure
(Figur-3)
Compare the structure of purines and
pyrimidines. Can you find out why the
distance between two polynucleotide
chains in DNA remains almost constant?
The proposition of a double helix
structure for DNA and its simplicity in
explaining the genetic implication became
revolutionary. Very soon, Francis Crick
proposed the Central dogma in molecular
biology, which states that the genetic
information flows from DNAàRNAàProtein.In some viruses the flow of information is in reverse direction, that is,
from RNA to DNA. Can you suggest a simple name to the process?